-Search query
-Search result
Showing all 18 items for (author: penzes & jj)

EMDB-41125: 
Zophobas morio black wasting virus strain NJ2-molitor virion structure
Method: single particle / : Penzes JJ, Kaelber JT
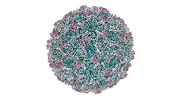
EMDB-41106: 
Zophobas morio black wasting virus strain UT-morio virion structure
Method: single particle / : Penzes JJ, Kaelber JT
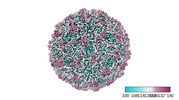
EMDB-41301: 
Zophobas morio black wasting virus strain OR-molitor virion structure
Method: single particle / : Penzes JJ, Kaelber JT
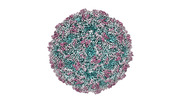
EMDB-41108: 
Zophobas morio black wasting virus strain UT-morio empty capsid structure
Method: single particle / : Penzes JJ, Kaelber JT
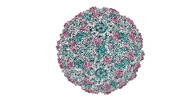
EMDB-41131: 
Zophobas morio black wasting virus strain OR-molitor empty capsid structure
Method: single particle / : Penzes JJ, Kaelber JT
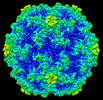
EMDB-28550: 
Acheta domesticus segmented densovirus, mature virion capsid structure
Method: single particle / : Penzes JJ, McKenna R, Tijssen P

EMDB-28553: 
Acheta domesticus segmented densovirus, high buoyancy (HB) capsid, a mixed population of empty and immature full particles
Method: single particle / : Penzes JJ, McKenna R, Tijssen P
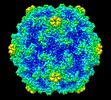
EMDB-28605: 
Acheta domesticus segmented densovirus high buoyancy fraction 1 (HB1) empty capsid structure
Method: single particle / : Penzes JJ, McKenna R, Tijssen P
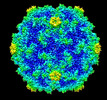
EMDB-28607: 
Acheta domesticus segmented densovirus VP-ORF1 virus-like particle
Method: single particle / : Penzes JJ, McKenna R, Tijssen P

EMDB-28604: 
Acheta domesticus segmented densovirus high buoyancy fraction 2 empty capsid structure
Method: single particle / : Penzes JJ, McKenna R, Tijssen P

EMDB-23986: 
Structure of the adeno-associated virus 9 capsid at pH 6.0
Method: single particle / : Penzes JJ, Chipman P, Bhattacharya N, Zeher A, Huang R, McKenna R, Agbandje-McKenna M

EMDB-23993: 
Structure of the adeno-associated virus 9 capsid at pH 5.5
Method: single particle / : Penzes JJ, Chipman P, Bhattacharya N, Zeher A, Huang R, McKenna R, Agbandje-McKenna M

EMDB-23999: 
Structure of the adeno-associated virus 9 capsid at pH 4.0
Method: single particle / : Penzes JJ, Chipman P, Bhattacharya N, Zeher A, Huang R, McKenna R, Agbandje-McKenna M

EMDB-24000: 
Structure of the adeno-associated virus 9 capsid at pH pH 7.4 in complex with terminal galactose
Method: single particle / : Penzes JJ, Chipman P, Bhattacharya N, Zeher A, Huang R, McKenna R, Agbandje-McKenna M

EMDB-24003: 
Structure of the adeno-associated virus 9 capsid at pH pH 5.5 in complex with terminal galactose
Method: single particle / : Penzes JJ, Chipman P, Bhattacharya N, Zeher A, Huang R, McKenna R, Agbandje-McKenna M

EMDB-23973: 
Structure of the adeno-associated virus 9 capsid at pH 7.4
Method: single particle / : Penzes JJ, Chipman P, Bhattacharya N, Zeher A, Huang R, McKenna R, Agbandje-McKenna M

EMDB-21667: 
Capsid structure of Penaeus monodon metallodensovirus at pH 8.2
Method: single particle / : Penzes JJ, Pham HT

EMDB-21668: 
Capsid structure of Penaeus monodon metallodensovirus following EDTA treatment
Method: single particle / : Penzes JJ, Pham HT
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
